Search tutorial
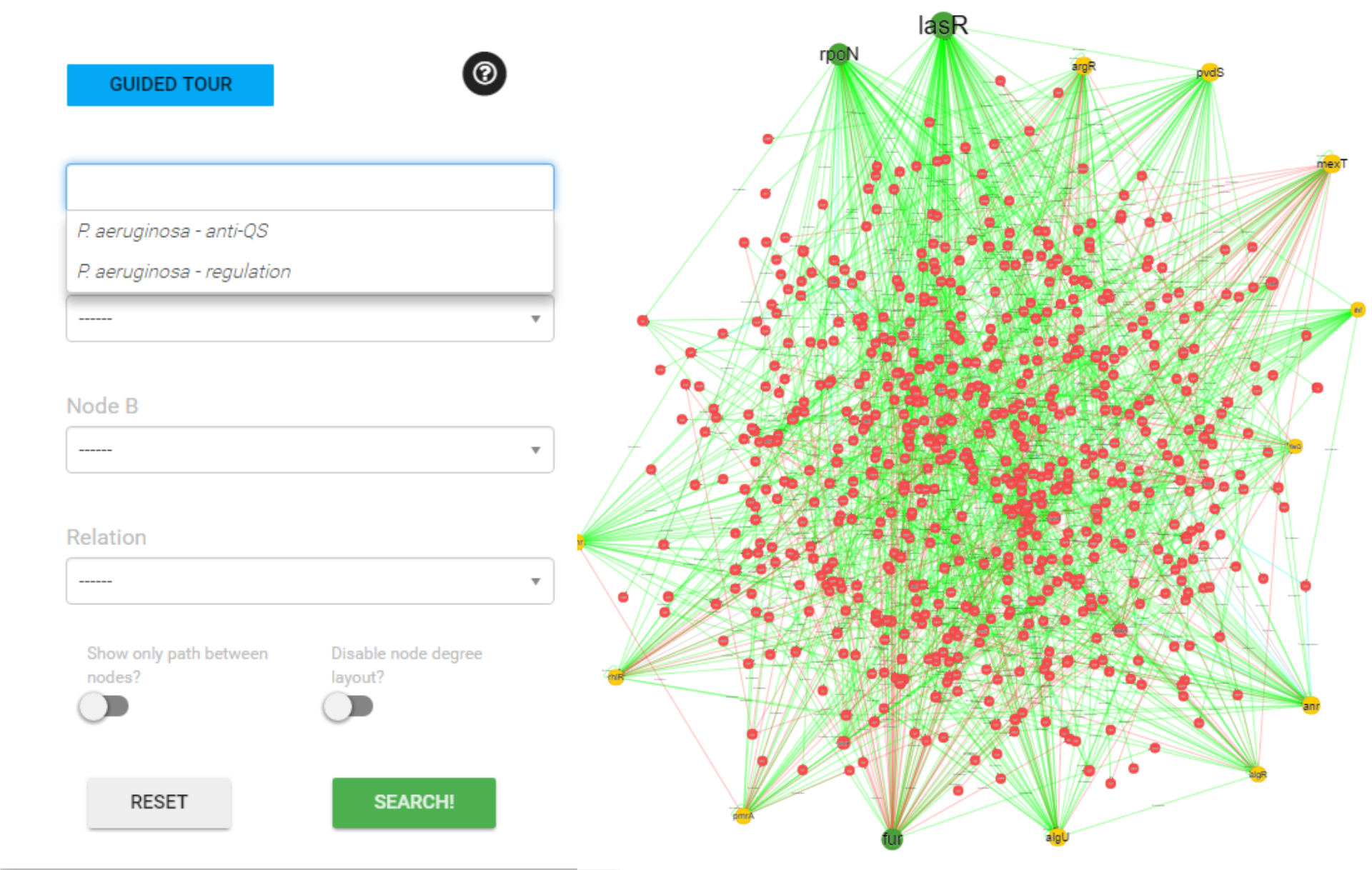
Search by Organism
Enter a organism name. The web will attempt to do auto-completion on the string you have entered based on the contents of the database.
If you select one of the auto-complete options, then when you submit the form you will be taken all polymicrobial agents in database related to this organism and their interactions.
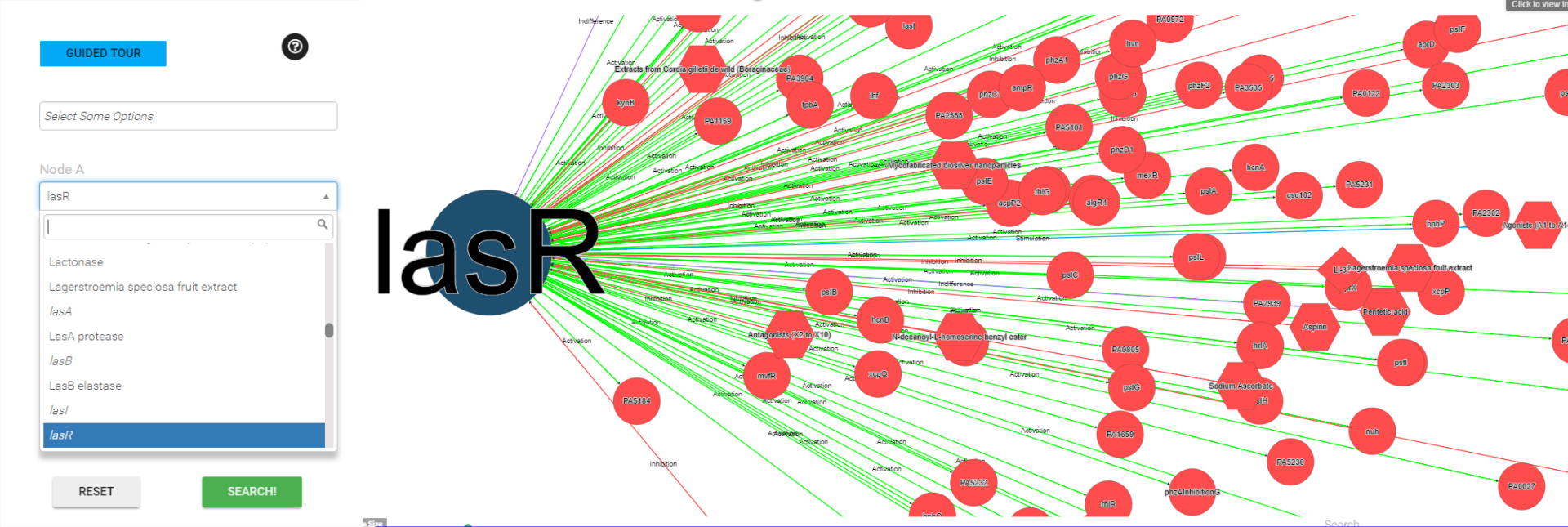
Search by polymicrobial agent
Enter a polymicrobial agent. The web will attempt to do auto-completion on the string you have entered based on the contents of the database.
If you select one of the auto-complete options, then when you submit the form you will be taken all polymicrobial agents in database that interact to this polymicrobial agent and their interactions.
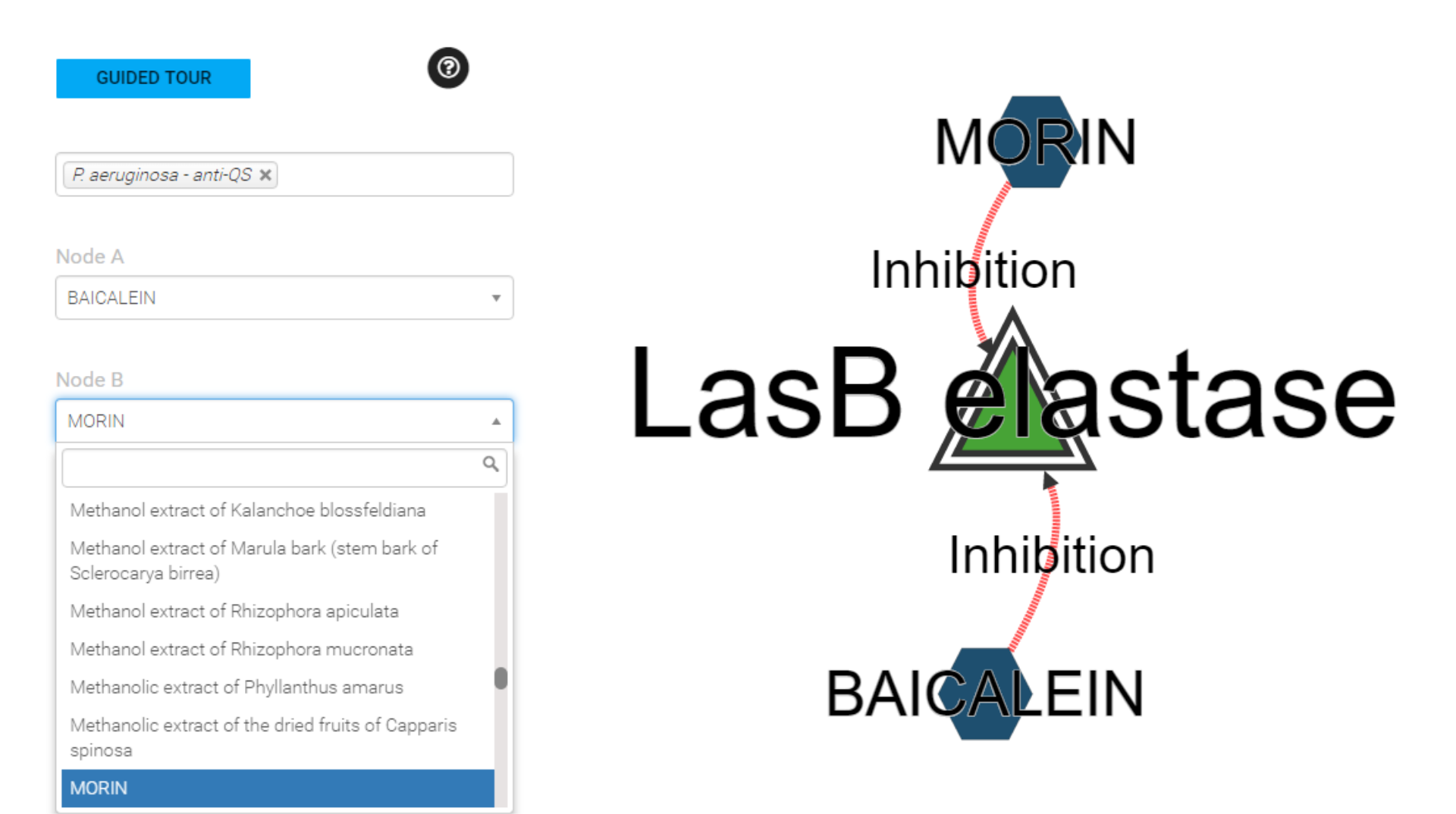
Search by polymicrobial combination
If you select two polymicrobial agents,
then when you submit the form the shortest path algorithm enables the identification of indirect relations, between polymicrobial agent A and polymicrobial agent B including intermediary agents and all the interactions documented for each agents..
Search by interaction type
Enter one polymicrobial interaction. The web will attempt to do auto-completion on the string you have entered based on the contents of the database.
If you select one of the auto-complete options, then when you submit the form you will be taken all polymicrobial interactions in database related of type selected.
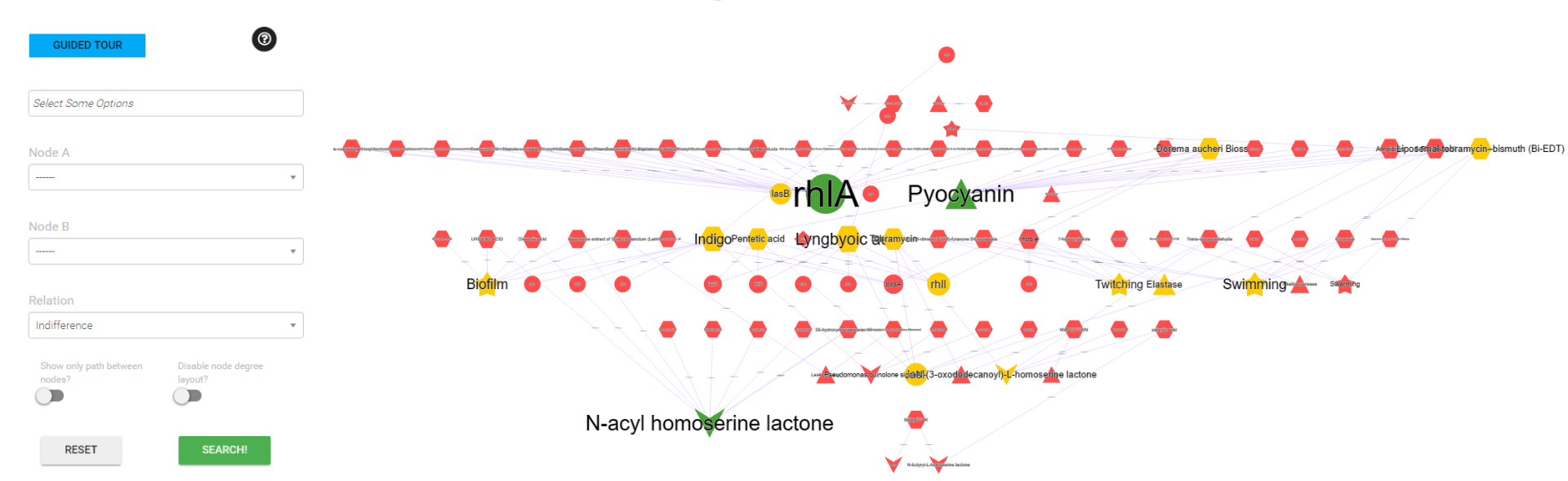
Search by Filter interaction
You can use only one option of form or You can combine all the different options for find more specific data, for example a specific combination (Ex. Activation/Inhibition) related to an type of network (Ex. P. aeruginosa - anti-QS)
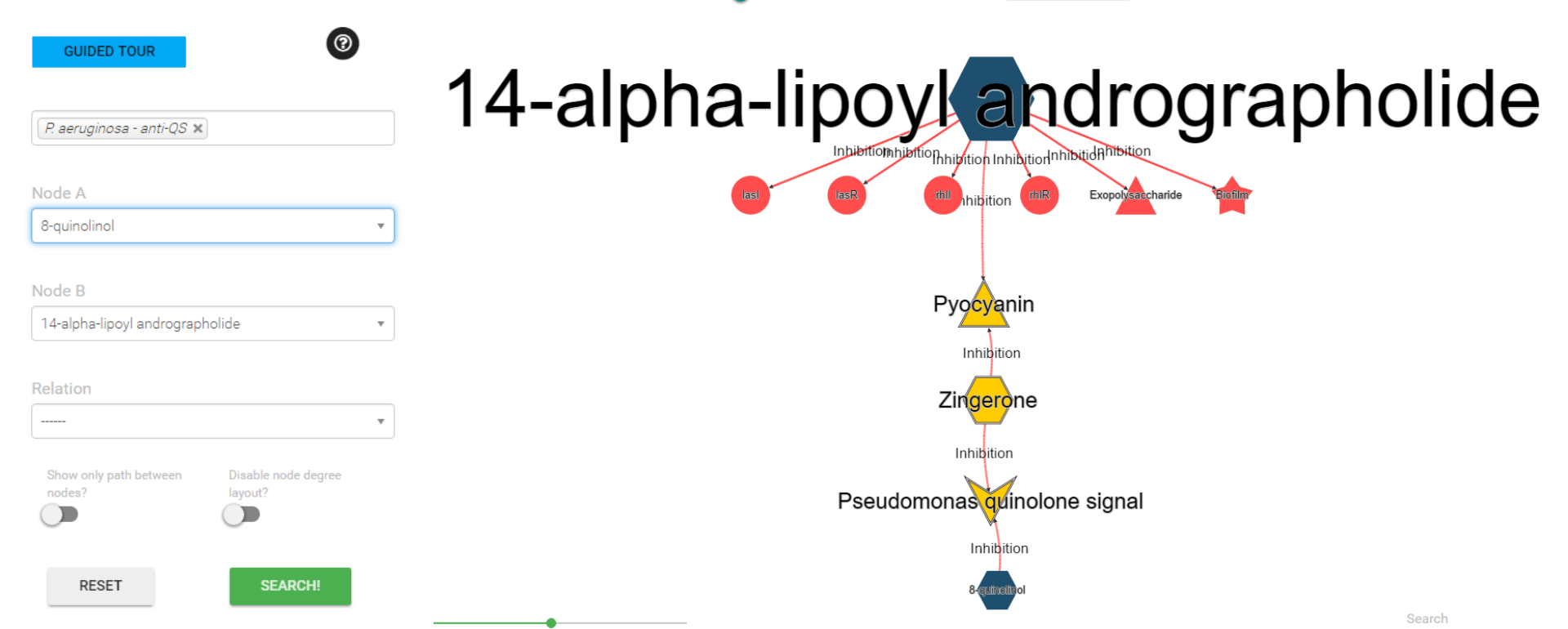
Search Interaction HeatMap
You can see the network of interactions of one polymicrobial gent from the heat map. Just click on the corresponding cell
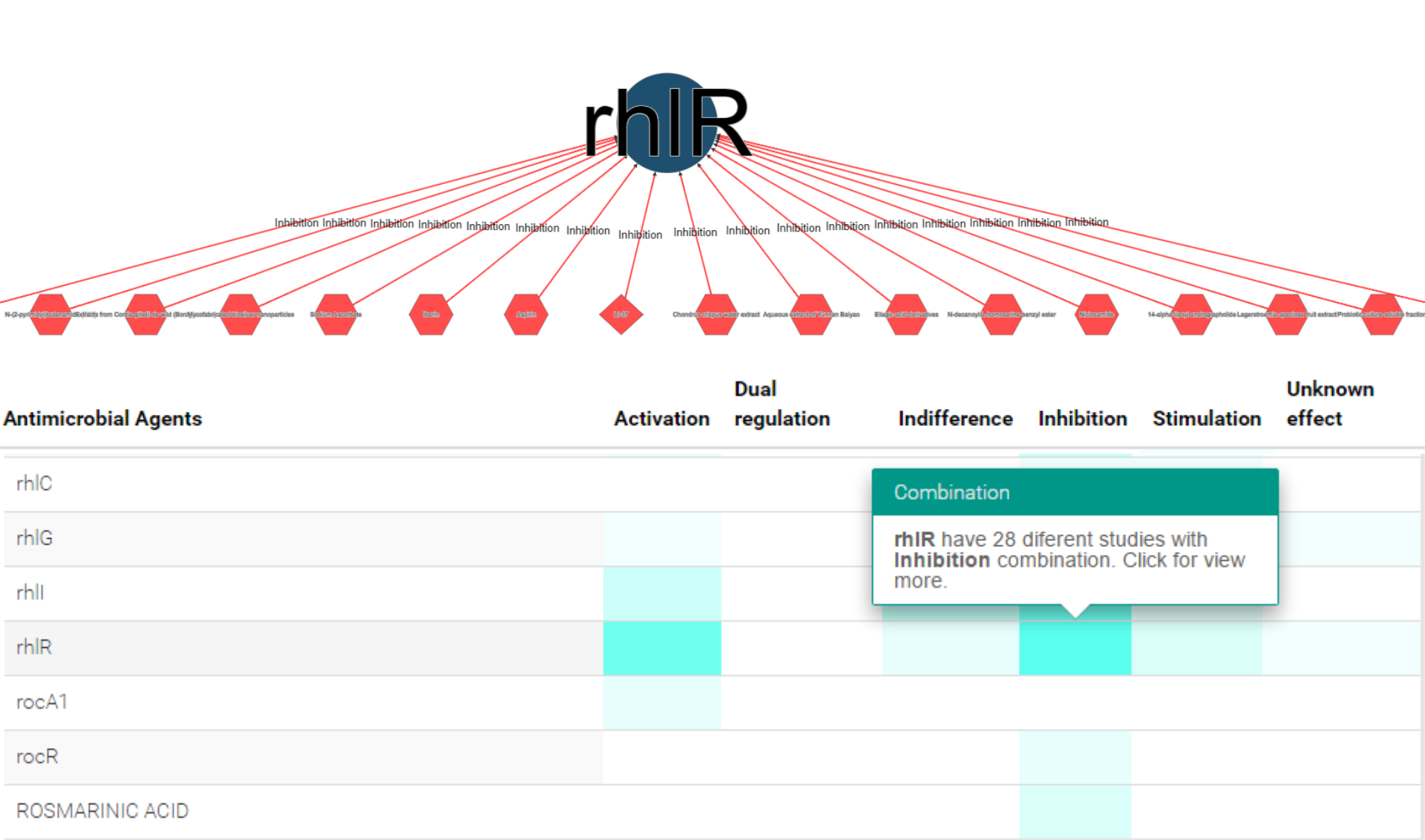
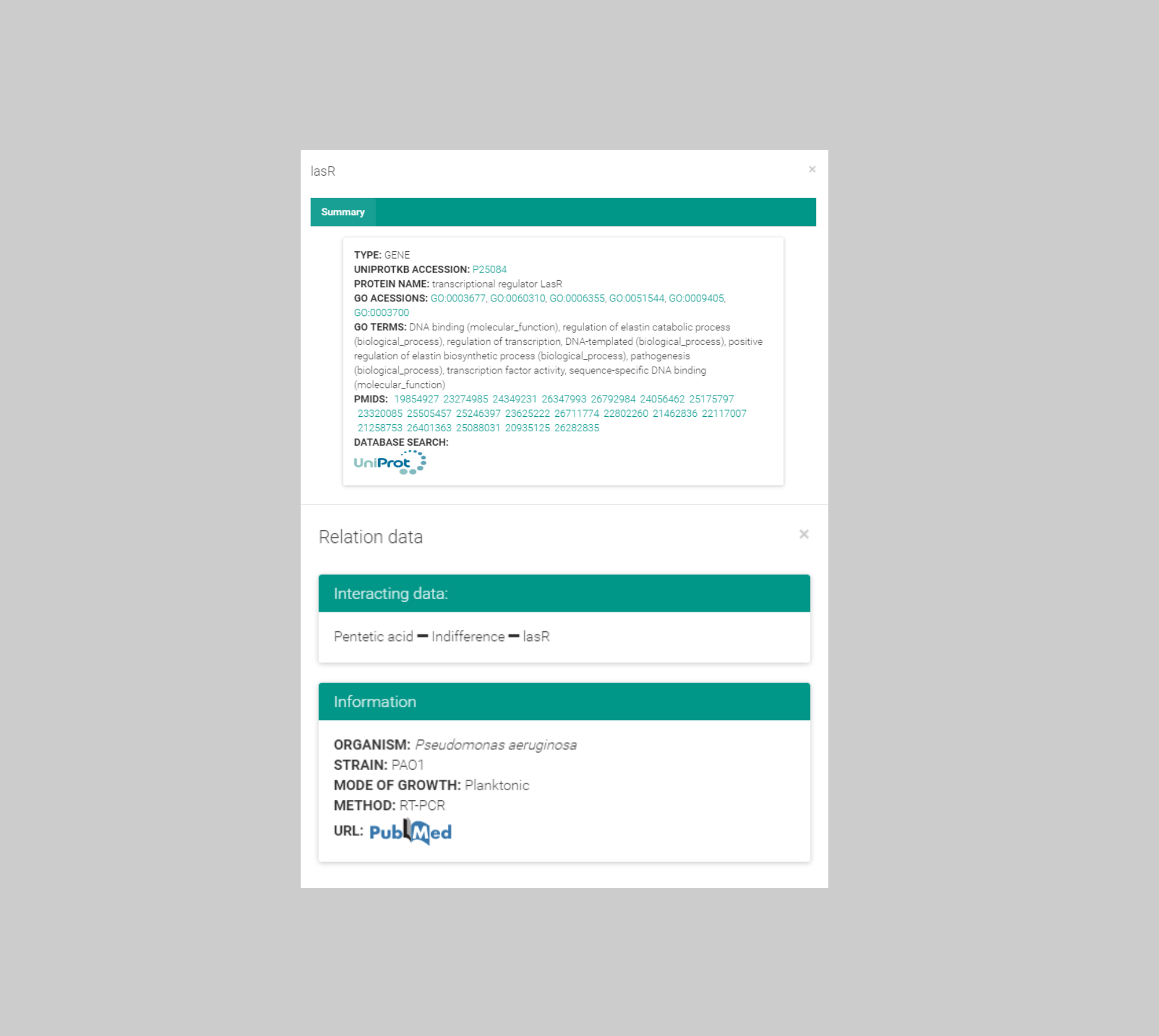
Antimicrobial agent & interaction information
Clicking at each node the website provides details on the represented polymicrobial agent, such as alternative names, chemical activity, cross-links to chemical and other external sources.
Moreover, the user may access the documents that supported the inclusion of the polymicrobial agent in the network,
both the original PubMed record and the curated abstract containing the expert revised annotations.
Likewise, each edge describes the nature of the documented combination and available susceptibility data.
Once again, the user may access the documents that supported the inclusion of the polymicrobial agent in the graph.